Computing trends in a climate grid with Tidyverse
Reading a NetCDF in the Tidyverse style!
Currently, it is possible to read a NetCDF file in a Tidyverse way through the package created by Elio Campitelli called metR. This library also contains many useful functions for visualizing and analyzing weather and climate data.
In this case, we will work with a daily grid of 5x5 km of the maximum temperature of the Balearic Islands (Spain) (Serrano-Notivoli 2019). To read it we will use a couple of functions from the metR package: GlanceNetCDF, to take a look at the variable name, dimensions, etc, and ReadNetCDF, to read the NetCDF file. Logically we will use the tidyverse package, which will allow us to organize the data and perform some simple calculations (remember that tidyverse is a collection of packages!), as well as the lubridate library, for the handling of dates, and the trend library to calculate the statistical significance of the trend of our data. We will also use the pals library to choose the color palette when we plot the results.
library(tidyverse)
library(lubridate)
library(metR)
library(trend)
library(pals) # color paletteIn order to work with the NetCDF data in the tidyverse style, we will specify that the output of the ReadNetCDF function is a data.frame:
# Download the Maximum Temperature (Tx) file for the Balearic Islands
# https://digital.csic.es/handle/10261/177655
# We extract information from the NetCDF (variable name, dimensions, etc)
file = "tmax_bal.nc"
variable <- GlanceNetCDF(file = file)
variable
## ----- Variables -----
## tx:
## Maximum temperature in Celsius degrees
## Dimensions: lon by lat by Time
##
##
## ----- Dimensions -----
## lon: 63 values from 0.970530815018679 to 4.55413081501868 degrees_east
## lat: 48 values from 38.3196292450381 to 40.4252292450381 degrees_north
## Time: 16071 values from 1971-01-02 to 2015-01-01
# Reading the NetCDF
tx <- ReadNetCDF(file = file,
vars = c(var = names(variable$vars[1])),# extraction of the name of the variable.
out = "data.frame") # output object
tx # voilà.
## Time lat lon var
## 1: 1971-01-02 40.42523 0.9705308 NA
## 2: 1971-01-02 40.42523 1.0283308 NA
## 3: 1971-01-02 40.42523 1.0861308 NA
## 4: 1971-01-02 40.42523 1.1439308 NA
## 5: 1971-01-02 40.42523 1.2017308 NA
## ---
## 48598700: 2015-01-01 38.31963 4.3229308 NA
## 48598701: 2015-01-01 38.31963 4.3807308 NA
## 48598702: 2015-01-01 38.31963 4.4385308 NA
## 48598703: 2015-01-01 38.31963 4.4963308 NA
## 48598704: 2015-01-01 38.31963 4.5541308 NAData management and handling
The objective is to aggregate the daily data on an annual scale. To do this we have to use several functions from the dplyr package, loaded in the tidyverse. First, however, we have to filter the NA values:
tx <- tx %>% filter(!is.na(var)) %>% as.tibble()
## Warning: `as.tibble()` is deprecated as of tibble 2.0.0.
## Please use `as_tibble()` instead.
## The signature and semantics have changed, see `?as_tibble`.
## This warning is displayed once every 8 hours.
## Call `lifecycle::last_warnings()` to see where this warning was generated.
tx
## # A tibble: 4,114,176 x 4
## Time lat lon var
## <dttm> <dbl> <dbl> <dbl>
## 1 1971-01-02 00:00:00 40.1 3.92 7.64
## 2 1971-01-02 00:00:00 40.1 3.98 7.69
## 3 1971-01-02 00:00:00 40.1 4.03 7.44
## 4 1971-01-02 00:00:00 40.1 4.09 8.29
## 5 1971-01-02 00:00:00 40.1 4.15 8.15
## 6 1971-01-02 00:00:00 40.1 4.21 8.35
## 7 1971-01-02 00:00:00 40.0 3.80 8.10
## 8 1971-01-02 00:00:00 40.0 3.86 8.04
## 9 1971-01-02 00:00:00 40.0 3.92 7.63
## 10 1971-01-02 00:00:00 40.0 3.98 7.70
## # ... with 4,114,166 more rowsOnce the NAs have been filtered, we proceed to carry out the annual aggregation at each grid point:
tx_annual <- tx %>%
mutate(Time = as_date(Time)) %>% # we convert the dates into lubridate format
mutate(year = year(Time)) %>% # We create the "year" column to perform the annual aggregate
group_by(year, lon, lat) %>% # we group by year, longitude and latitude
summarise(annual_tx = mean(var)) %>% # computing yearly mean
ungroup()
## `summarise()` regrouping output by 'year', 'lon' (override with `.groups` argument)
tx_annual
## # A tibble: 11,520 x 4
## year lon lat annual_tx
## <dbl> <dbl> <dbl> <dbl>
## 1 1971 1.20 38.9 20.8
## 2 1971 1.20 38.9 20.9
## 3 1971 1.26 38.9 20.2
## 4 1971 1.26 38.9 20.3
## 5 1971 1.26 38.9 20.8
## 6 1971 1.32 38.9 20.7
## 7 1971 1.32 38.9 20.6
## 8 1971 1.32 38.9 20.7
## 9 1971 1.32 39.0 21.0
## 10 1971 1.32 39.0 20.3
## # ... with 11,510 more rowsAs we can see, the average maximum temperature for each year and grid point is shown.
Trend Computation and its statistical significance
In this way, we can already calculate the annual trend and its statistical significance with the trend package.
To do this we will use the sens.slope function to calculate the slope of the trend (increase/decrease per year); and the mk.test function (Mann test Kendall), to verify if the trend is statistically significant or if it is the result of the variability of the thermometric series itself. Furthermore, as this is a non-parametric test, our data will not have to fit a Gaussian distribution.
tx_trend <- tx_annual %>%
group_by(lon,lat) %>% # we group by lon and lat to perform the calculation in each cell
summarise(slope = sens.slope(annual_tx)$estimates *10,
sign = mk.test(annual_tx)$p.value)
## `summarise()` regrouping output by 'lon' (override with `.groups` argument)
tx_trend
## # A tibble: 256 x 4
## # Groups: lon [38]
## lon lat slope sign
## <dbl> <dbl> <dbl> <dbl>
## 1 1.20 38.9 0.122 0.156
## 2 1.20 38.9 0.122 0.156
## 3 1.26 38.9 0.110 0.115
## 4 1.26 38.9 0.112 0.120
## 5 1.26 38.9 0.147 0.0834
## 6 1.32 38.9 0.136 0.0983
## 7 1.32 38.9 0.133 0.111
## 8 1.32 38.9 0.146 0.0869
## 9 1.32 39.0 0.150 0.0645
## 10 1.32 39.0 0.133 0.0983
## # ... with 246 more rowsThe value of the Sen’s slope gives us the rate of change per 10 years (we have multiplied by 10), as well as the statistical significance -provided for the Mann-Kendall test- of the trend indicated through the p-value. Now we will visualize the results indicating those cells that show a statistical significance to the increase or decrease of the maximum annual temperature.
ggplot()+
geom_tile(data = tx_trend, aes(x = lon, y = lat, fill = slope))+
scale_fill_gradientn(colors = rev(pals::linearlhot(100)), name = "ºC/10y", limits = c(0.1,0.45)) +
geom_point(data = filter(tx_trend, sign < 0.01),aes(x = lon, y = lat, color = "Sign. trend \n p-value <0.01"),
size = 0.4, show.legend = T) +
scale_color_manual(values = c("black"), name = "")+
coord_fixed(1.3)+
xlab("Longitude") + ylab("Latitude")+
labs(title = "Decadal trend of maximum temperature in the Balearic Islands",
subtitle = "(1971-2014)",
caption = "Database: STEAD (Serrano-Notivoli,2019)")+
theme_bw() +
guides(fill = guide_colourbar(barwidth = 9, barheight = 0.5, title.position="right"))+
theme(legend.position = "bottom")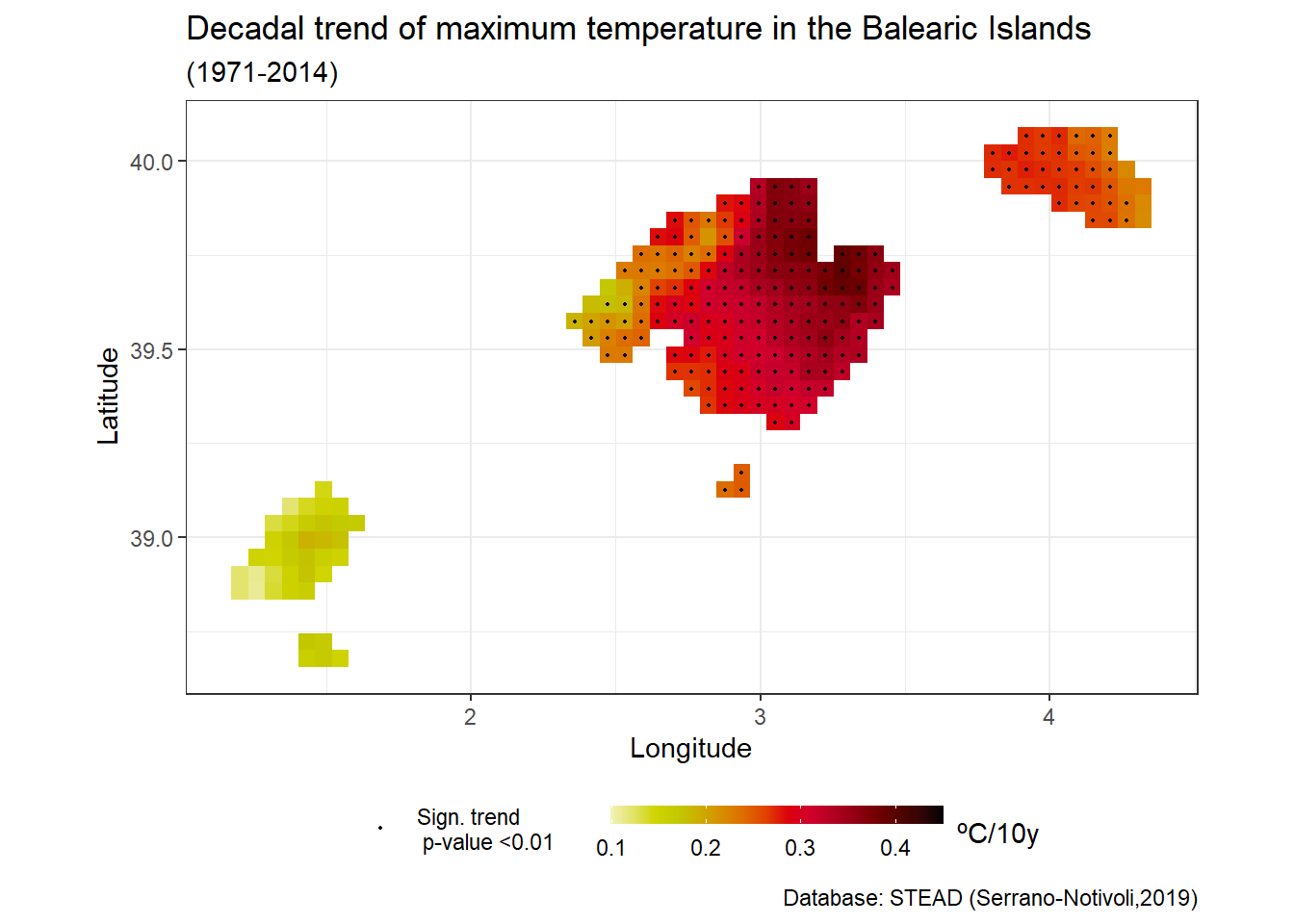
If you have any questions, please do not hesitate to leave a comment.
Greetings!